In the complex ecosystem of the gut, where billions of microorganisms coexist, the presence of an unexpected aid can sometimes spark interesting changes. Specifically, T. mu, otherwise known as Tritrichomonas musculis, had a presence that triggered a series of unexpected transformations within the host’s microbiome.
Image courtesy of Dr. Chudnovskiy Merad & Dr. Arthur Mortha
A New Hypothesis
For years, immunologists have known that the gut microbiome, especially bacteria, plays a crucial role in training our immune system. One of its key outputs is Immunoglobulin A (IgA), an antibody that coats the gut lining and neutralizes threats before they can breach the walls… but what about protozoa?
A recent study published by researchers at the University of Toronto proposed an interesting question: Could a protozoan like T. mu influence the production of IgA, and if so, how?
The Experiment
To find the answer to that question, researchers introduced T. mu into the guts of mice and waited. Three weeks later, they checked the mice’s immune systems, and once graphed, the results were quite notable:
- IgA levels in the blood and gut had surged.
- The number of IgA-producing plasma cells had increased dramatically.
- Specialized helper T-cells, also known as T-follicular helper (Tfh) cells, had multiplied in the gut’s lymphoid tissues.
Somehow, T. mu managed to turn on IgA production and amplify it. To understand how this was happening, the team looked closer. They discovered that T. mu was activating a specific immune pathway involving a molecule called ICOS. This molecule acts like an agreement between T-cells, which wipe out invaders, and B-cells, which create antibodies, helping them coordinate the production of high-quality antibodies.
When the researchers blocked ICOS, the IgA surge disappeared. When they removed the T-cells entirely, the effect vanished too. Turns out, T. mu was creating a specific T-cell dependent immune response. To see how this affected the gut, the team used 16S rDNA sequencing, a method that identifies bacteria by reading a genetic code found in all bacterial species.
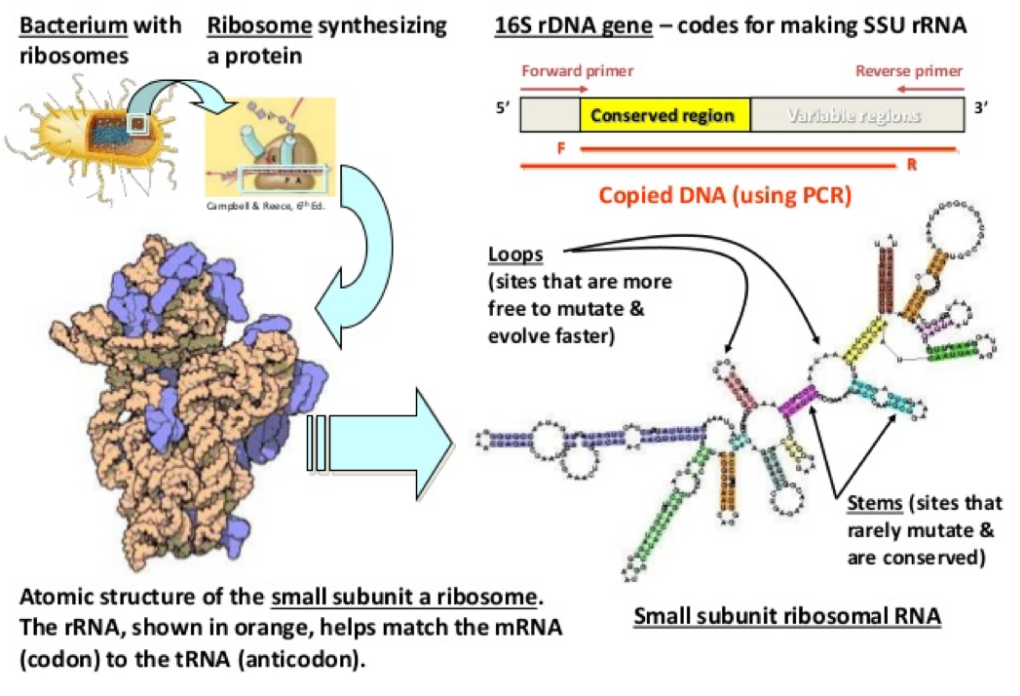
Image courtesy of the Fankhouser Lab
With this method, they discovered that T. mu colonization caused a clear shift in the types of bacteria being targeted by IgA. Specifically, bacteria coated with IgA looked different from those that weren’t, suggesting that T. mu helped the immune system refine its focus. T. mu was making more antibodies and helping the body decide where to send them.
Broader Impact
The researchers also found that the IgA produced in response to T. mu was more targeted. It coated gut bacteria more efficiently and recognized a broader range of microbial proteins. In other words, T. mu was increasing the quantity of IgA and improving its quality.
A more refined IgA response can help the body distinguish between invaders, reducing unnecessary inflammation and enhancing protection against real threats. Even more surprisingly, the effects of T. mu‘s colonization lasted for over a year, which means the immune system remembered its response for twelve months after the protozoan settled.
Reimagining the Microbiome
When we think about microbes that help our immune system, most people think of bacteria. But now, T. mu shows that protozoa can also act as “natural adjuvants,” enhancing the immune responses without causing harm.
What Next?
In this case, the implications are many. Could protozoa be harnessed to improve oral vaccines? Could they help module immune responses in conditions like food allergies or inflammatory bowl disease? Could we, one day, design probiotics that include beneficial bacterial protozoa and not just bacteria?
We don’t have all the answers yet, but thanks to this study, we’re asking better questions about the extent of microbes in our microbiome.
Citations
Cao, E. Y., Burrows, K., Chiaranunt, P., Popovic, A., Zhou, X., Xie, C., Thakur, A., Britton, G., Spindler, M., Ngai, L., Tai, S. L., Dasoveanu, D. C., Nguyen, A., Faith, J. J., Parkinson, J., Gommerman, J. L., & Mortha, A. (2024). The protozoan commensal Tritrichomonas musculis is a natural adjuvant for mucosal IgA. The Journal of experimental medicine, 221(12), e20221727. https://doi.org/10.1084/jem.20221727.
Woo, P. C., Lau, S. K., Teng, J. L., Tse, H., & Yuen, K. Y. (2008). Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases, 14(10), 908–934. https://doi.org/10.1111/j.1469-0691.2008.02070.x.
No AI was used in the writing or editing of this material.